

VMD state contains all the information needed to start a new VMD sessionįrom it, without losing what you have done.ġ In the VMD Main window, choose the File Save State menu item. The image that you have created using VMD can be saved,Īlong with all representations you have created, as a VMD state. This is especially useful forĤ Using the shift key while pressing the mouse button allows you to Use the right buttonģ Using the Zoom controls (f) you can display the entire In addition, the highlighted residue willĪppear in your OpenGL Display window in yellow and bondĭrawing method, so you can visualize it easily. 9(a) with a list of the aminoĪcids (e) and their properties (b)&(c) will appear in your screen.Ģ With the mouse, click over different residues (e) in the list and TheĮxtension allows you to pick and display one or more residuesġ Choose the Extensions Analysis Sequence Viewer menu item.Ī window Fig.

Useful to find and display different amino acids quickly. When dealing with a protein for the first time, it is very Of this section, the Graphical Representations window Representations you have created and modify each one independently.Īlso, you can switch each one on/off by double-clicking on it orĭelete each one by using the Delete Rep button (b). For this last representationĦ Note, that with the mouse, you can select the different This selects all the water molecules that are within a distance of 3įinal representation by pressing again the Create Rep button.Ĭoloring Method Molecule and type protein in the (in fact only the oxygens) present in our system.ħ In order to see which water molecules are closer to the protein Screen you will be able to see the different Lysines and Glycines.Ītoms text entry type water.
Play a fundamental role in the configuration of polyubiquitin chains.ĥ Now, change the current representation's Drawing Method to CPK styleĪnd the Coloring Method to ResID in the Draw Style tab. Protein by typing (resname LYS)or(resname GLY). You can see properties that can be used to select parts of a Try to display sheets instead of helices by typing the appropriateĬombinations of boolean operators can also be usedģ In order to see the molecule without helices and sheets, type theįollowing in Selected Atoms: (not helix)and(not betasheet) Will find a list of possible selections you can type. VMD will show just the helices present in our molecule.Ģ In the Graphical Representations window choose Type helix and press the Apply button or hit the Enter/Return key on your keyboard. Let's look at different independent (and interesting) parts ofġ In the Selected Atoms text entry Fig. This allows you to distinguish non-polar residues (white),īasic residues (blue), acidic residues (red) and polar residues (green).Ģ Select Coloring Method Structure (c) andĬonfirm that the NewCartoon representation displays colors consistent with secondary Way you can see more easily the volumetric distribution of the protein.ġ Now, let's modify the colors of our representation. Each atom is now represented by a sphere. The Thickness of the lines by using the controlsĤ Now, choose VDW(van der Waals) from Drawing Method. The drawing style (the default is Lines).ģ Each Drawing Method has its own parameters. Graphical representation used to display your molecule.Īnd color (c) of the representation. A window called Graphical Representations will appear and You to identify different structures in the protein.ġ Choose the Graphics Representations. Here, we will explore those that can help VMD can display your molecule using a wide variety ofĭrawing styles. Your molecule moves around the point you have selected. It allows you to specify the point around which rotations are done.Ħ Select the Center menu item and pick one atom at one of theĮnds of the protein The cursor should display a cross.ħ Now, press r, rotate the molecule with the mouse and see how In order to actually load the file you have to press Load
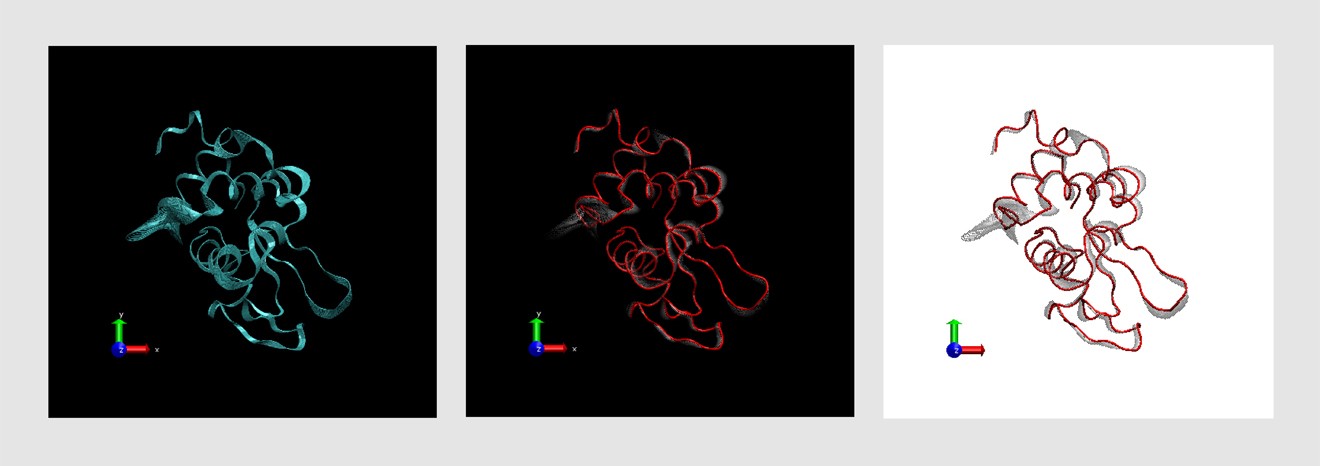
The file, you will be back in the Molecule File Browser window. Another window, the Moleculeįile Browser (b), will appear in your screen.ġUBQ.pdb in vmd-tutorial-files. A pdbġUBQ.pdb, that contains the atom coordinates of ubiquitinġ Choose the File New Molecule. To look for interesting structural properties of proteins using VMD. In this unit you will build a nice image of ubiquitin whileīecoming accustomed to basic VMD commands. Next: Multiple Molecules and Scripting Up: VMD Tutorial Previous: Introduction


 0 kommentar(er)
0 kommentar(er)
